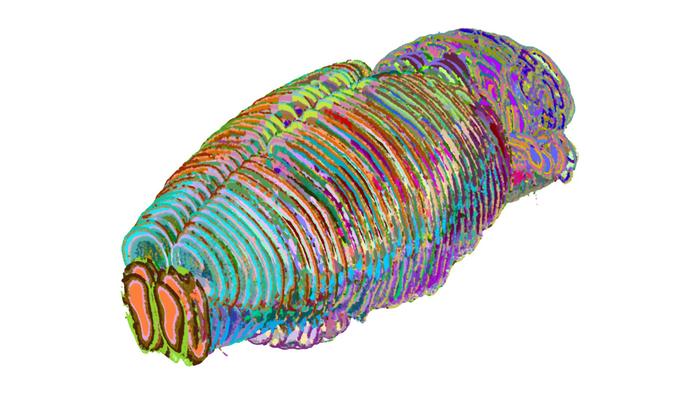
Researchers at the University of California, San Francisco and Allen Institute have used AI to create one of the most detailed maps of the mouse brain, featuring 1,300 regions and subregions, including previously uncharted areas.
The AI model, called CellTransformer, automatically identifies brain subregions from large-scale spatial transcriptomics data. The findings, published in Nature Communications, provide an unprecedented level of detail that allows researchers to link specific functions, behaviours and disease states to smaller, more precise cellular regions.
“It’s like going from a map showing only continents and countries to one showing states and cities,” said Bosiljka Tasic, director of molecular genetics at the Allen Institute and one of the study authors. “This new, detailed brain parcellation solely based on data, and not human expert annotation, reveals previously uncharted subregions of the mouse brain.”
CellTransformer uses the same transformer framework as ChatGPT to analyse relationships between nearby cells in space. The model predicts a cell’s molecular features based on its local neighbourhood, building up a detailed map of tissue organisation.

“Our model is built on the same powerful technology as AI tools like ChatGPT. Both are built on a ‘transformer’ framework which excels at understanding context,” said Reza Abbasi-Asl, associate professor of neurology and bioengineering at UCSF and senior author of the study.
The model successfully replicates known brain regions such as the hippocampus, but also discovers previously uncatalogued subregions in poorly understood areas like the midbrain reticular nucleus.
Unlike previous brain maps, CellTransformer’s boundaries are defined by cellular and molecular data rather than human interpretation. The Allen Institute’s Common Coordinate Framework served as the validation standard for the model’s accuracy.
CellTransformer’s capabilities extend beyond neuroscience and can be applied to other organ systems and tissues, including cancerous tissue, where spatial transcriptomics data is available.